Mice are very different from humans — but when it comes to medical research, sometimes they’re all we’ve got.
Assistant Professor Shai Shen-Orr of the Technion’s Rappaport Faculty of Medicine and Technion Ph.D. candidate Rachelly Normand, together with Professor Rob Tibshirani and Ph.D. candidate Wenfei Du from Stanford University, have developed a revolutionary, big-data-driven, machine learning algorithm that can predict the results of human gene expression based on preliminary results of mouse studies.
The tool could speed up the development of new medical therapies and may even reduce future experiments done on mice.
Why Mice?
Mouse model studies are critical in experiments that cannot be performed in humans due to ethical considerations. This is particularly true of studies involving diseases and physiological processes in the brain, spleen, and heart, or in testing the efficacy of new treatments for certain diseases.
Despite this, it’s not always easy to translate findings in mice to humans. Because of our physiological, genetic, life expectancy, and environmental differences, a drug or treatment that works for mice may fail when tested in humans.
From Raw Data to Big Insights
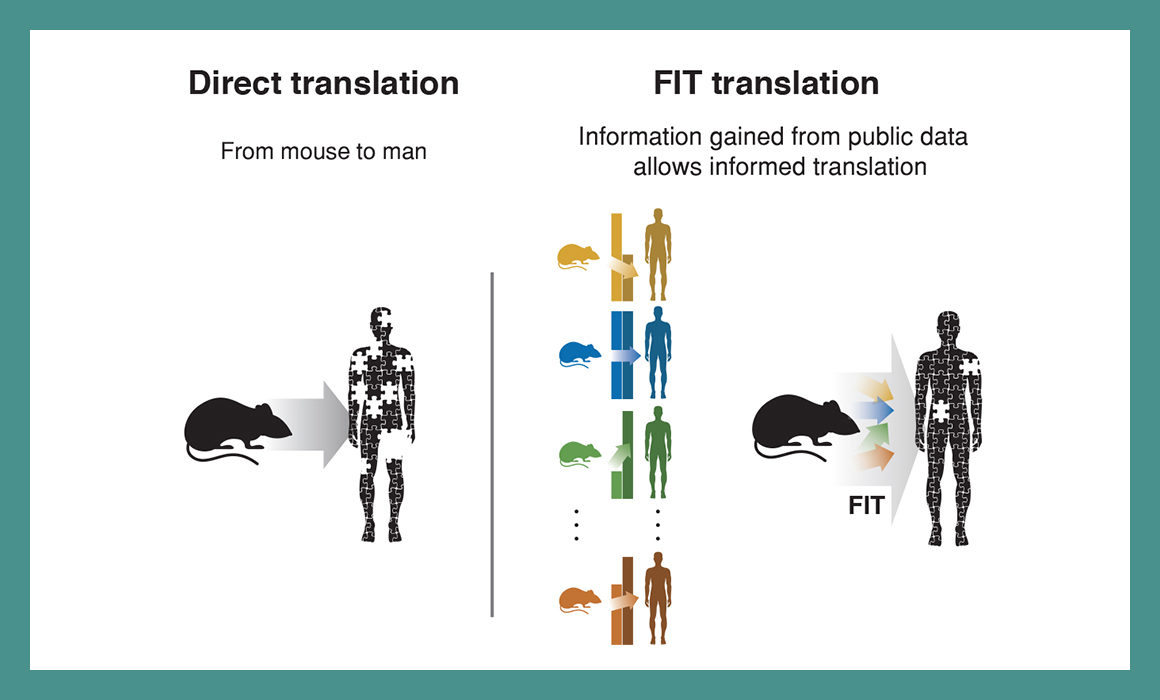
The new tool developed at the Technion predicts how relevant mouse test results will be to human physiology — speeding up the development of new drugs and dramatically reducing the cost.
It was made possible by a relatively new norm in research: uploading raw data from scientific studies to the internet. This practice began with the human genome project, but has evolved and grown substantially; there are now measurements of more than 2 million samples online, mostly from tissues of human patients and disease animal models.
Most of this data is not used beyond the study in which it was generated. But the researchers believed that data held hidden treasures — they just had to extract them.
Found in Translation
The researchers measured the levels of mRNA, a central component in protein production, to better understand the relationship between gene expression in mice and an equivalent human condition. Then, they developed an algorithm that translated experiments conducted on mice, enabling researchers to better extrapolate the implications on humans.
The system is called Found in Translation (a play on “lost in translation”), or FIT. It is able to identify whether information collected from prior studies could be relevant for a new animal study. If the information is relevant, the system adjusts the results measured in the new study, allowing researchers to understand how the new study findings in mice are relevant to humans.
The Power of FIT
The researchers used 170 different mouse studies to evaluate FIT’s performance. In 88% of cases where FIT is predicted to be relevant to the new mouse experiment, FIT is able to correctly “translate” between mice and humans. This improves our ability to infer results between species by 50%.
Additionally, researchers tested FIT’s predictive powers in a Crohn’s disease mouse model. FIT predicted that the ILF3 gene would be expressed in humans, even though it is not expressed in mice. In a validation experiment, researchers found that the protein product of the ILF3 gene is indeed expressed in Crohn’s patient samples — a completely new discovery that would not have been discovered without the algorithm!
By bridging the “cross-species gap,” FIT is not only translating animal model findings into human insights, but it’s also improving research accuracy, preventing false leads, and shortening drug and therapy development. It’s an incredible win for researchers everywhere, and it’s thanks to the brilliant minds at the Technion.